Rushing Remediation
Field studies have shown that many organic contaminants degrade naturally by microorganisms present in soil and groundwater. This naturally occurring bioremediation, known as natural attenuation, has been accepted as a passive remediation process for contaminated soil and groundwater. When the natural degradation is insufficient to meet remediation goals protective of human health and the environment, there may be attempts to increase the degradation rate by adding nutrients to stimulate the growth of the native microbial populations.
Active bioremediation, which includes adding specialty microbes, may mimic the naturally occurring process, but it has the advantage of taking the guesswork out of nutrient-stimulated bioremediation. For example, the degradation rate of the target compound is known and the specialty microbes can be delivered in populations higher than a stimulated natural system can achieve. In addition, adding specialty microbes can be faster and less expensive than attempts to stimulate indigenous microbes.
The Genetic Basis of Cometabolism
Cometabolic aerobic oxidation is the microbial breakdown of a contaminant in which the contaminant is oxidized incidentially by an enzyme produced during the metabolism of another compound.1 In other words, a cometabolite is an enzyme produced by microbiological metabolism that aids in the degradation of a contaminant. Cometabolism in Pseudomonas bacteria is possible because they have genetic coding available for metabolic pathways, which produce enzymes. The development and preservation of these genetic traits may be indirect evidence that the cometabolism provides some advantage to the bacteria. This characteristic of Pseudomonads provides an effective mechanism for in situ bioremediation of a wide range of organic contaminants.
Cometabolic aerobic oxidation is the microbial breakdown of a contaminant in which the contaminant is oxidized incidentally by an enzyme produced during the metabolism of another compound. |
Pseudomonas bacteria are well known for their ability to break down a wide range of chemicals.2 Their natural ability to tolerate and degrade contaminants can be enhanced by selective regeneration in the laboratory. At times the combination of several strains of microbes in a synergistic consortium can induce further microbial capabilities.3
Pseudomonads represent the most widely studied bacterial genera for investigations involving the degradation of complex hydrocarbons. Pseudomonads are gram negative aerobic rods that are ubiquitous in nature. Early studies in 1926 in the Netherlands by den Doorende Jong demonstrated that many different organic compounds supported the growth of Pseudomonads.4 These early studies led to many investigations in countless laboratories around the world into the complex biochemical pathways that were discovered to be present in these bacteria. As our ability to understand the genetic basis for this metabolism grew in the 1970s more laboratories devoted time to understanding the genes encoding for this metabolism.
Since the 1960s certain investigators ( I. C. Gunsalus, D. T. Gibson, H. J. Knackmuss, B. W. Holloway, A. M. Chabrabarty, R. H. Olsen, K. N. Timmis, J. C. Spain, L.P. Wackett and G. J. Zylstra) have been associated with studies involving the molecular basis for the degradative metabolism of the Pseudomonads.5 Through their scientific efforts to understand the molecular basis of the degradative metabolism of Pseudomonads, the successful application of the strains into the environment has been possible.
One of the advantages of Pseudomonads is their aerobic respiration. The fact that aerobic oxidation is possible broadens the scope of applications. Pseudomonads are suitable for use in not only aerobic soil and groundwater applications, they have been used in applications open to the atmosphere and in combination with air sparging.
Accelerating Site Closure by Cometabolism
A case study shows that while the naturally occurring microbial population may have partially degraded contaminants, the degradation was slow and incomplete. Using specialty microbes, the degradation was completed in an efficient time frame.
Pseudomonas bacteria are well known for their ability to break down a wide range of chemicals. |
At a former chemical mixing company, contamination by a wide range of volatile organic chemicals was found in soil and groundwater. The contamination resulted from the accumulation of years of incidental spills. The volume of contaminated soil within the source area was estimated to be 2,200 tons. The soil contaminants were mainly tetrachlororethene (PCE) and trichloroethene (TCE), and 1,1,1-trichloroethane (1,1,1-TCA).
The size of groundwater plume that resulted from leaching soil contaminants was estimated to be 12,000 square feet. The affected aquifer was a perched sandy gravel zone approximately four feet thick. The main groundwater contaminants were 1,1,1-trichloroethane (1,1,1-TCA), 1,1-dichloroethane (1,1-DCA), vinyl chloride and 1,2-dichloroethene (1,2-DCE). There may have been some natural degradation as evidenced by the relatively high concentration of cis-1,2-DCE and lower concentrations of PCE and TCE in groundwater in comparison with the ratios found in soil.
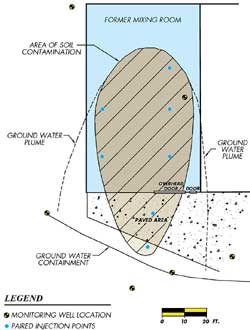
After the nature and extent of contamination was determined, the property owners found that remediation options were beyond their financial means. The owners' only option was to install a containment system to prevent further migration of contamination. A pump and treat containment system was installed at the property line to control the groundwater contamination in 1993. Figure 1 (opening picture on this page) shows the property layout, including the soil source area and groundwater plume limits.
During the operation of the pump and treat system the groundwater concentrations were monitored to verify the contamination was controlled. The periodic monitoring tracked the contaminant levels as time passed. After eight years, several changes were observed in the contaminant concentrations. Of the nine compounds detected in groundwater the greatest amount of degradation was in TCE and 1,1,1-TCA. The concentrations of both of these compounds decreased by about 50 percent. Meanwhile, the PCE and vinyl chloride concentrations increased suggesting both continued leaching from the source area and partial breakdown to vinyl chloride. The total VOC mass increased by about 25 percent during the pump and treat operation, indicating that whatever degradation was taking place it did not keep up with the leaching rate.
In 2001, the property was sold to a new owner who wanted to develop the site and was willing to remediate the source area and terminate the obligation to provide groundwater containment. Cl-out microbes were injected in the source area on a monthly basis for six months. Cl-out microbes are a specialty blend of naturally-occuring microbes provided by CL Solutions for the bioremediation of chlorinated solvents. Cl-out microbes are Pseudomonas bacteria that cometabolize a wide range of chlorinated aliphatic compound in the presence of dextrose.
Six injection points were installed in the contaminated soil area (Figure 1). Since the source of contamination was suspected to be surface spills, the injection points were installed in nests to increase the distribution through the soil profile. At each nested pair, one injection screen was set near the surface of the soil (one to two feet deep) and a second screen was set in the water-bearing formation at nine to 12 feet deep.
During each inoculation, 275 to 385 gallons of inoculant was injected into the subsurface. The inoculant was split among the injection points so that 40 to 55 gallons of solution was injected into each pair of injection points.
Pseudomonads are suitable for use in not only aerobic soil and groundwater applications, they have been used in applications open to the atmosphere and in combination with air sparging. |
Inoculations were made on a 30-day schedule to maintain the high microbial population for six months. The 30-day inoculation schedule was based on observations at other sites where the injected microbial population diminished to less than optimum levels in approximately 30 days. Microbial plate counts performed on groundwater samples showed that the population diminished even more rapidly at this site. Population counts taken 30 days after injections showed that the population of injected microbes was less than 10,000 colony forming units per milliliters (cfu/ml). The effective population is above 1,000,000 cfu/ml.
The biological activity indicators prior to bioremediation showed the groundwater environment was aerobic. The dissolved oxygen (DO) concentration prior to the first inoculation was 2.63 milligrams per liter (mg/l). During the inoculation the DO concentration decreased steadily to as low as 1.0 mg/l. The groundwater oxygen-reduction potential (ORP) was 115 prior to initiating the bioremediation but decreased to less than -100 after the first three inoculations. The Cl-out population appeared to have been suppressed by the consumption of the dissolved oxygen more rapidly than it was replenished by infiltration.
Contaminant tracking showed that there was immediate degradation of contamination. After the first inoculation, the concentrations of TCE and PCE were reduced by more than 50 percent. The concentration of 1,1,1-TCA and cis-1,2-DCE were also reduced significantly. After eight months nearly all of the target compounds were below drinking water standards. Only vinyl chloride and cis-1,2-DCE were above drinking water levels.
The last inoculation was made in November 2001. The post-closure monitoring showed that the contaminant concentrations continued to decrease through March 2002. The site will be closed after two years of post closure monitoring verifies that the cleanup goals have been achieved and maintained.
The combined total cost to remediate both soil and groundwater was approximately $80,000. This total includes the installation of injection wells, monthly application of bioremediation for eight months, and groundwater monitoring during bioremediation. The unit costs based on the volumes treated are $36.00 per ton of soil or $0.76 per gallon of groundwater.
Conclusion
Active bioremediation can be used cost effectively to accelerate site remediation. The addition of microbes selected for their ability to degrade target chemicals can improve the natural degradation that may be taking place, bringing a site to closure many years sooner than would be possible with enhanced natural attenuation. In the case study the site remediation was accelerated to restore the value of the property and make redevelopment possible.
References
1. U.S. Environmental Protection Agency. (July 2000). Engineered Approaches to In Situ Bioremediation of Chlorinated Solvents; Fundamentals and Field Applications (EPA 542-R-00-008). Washington, D.C.: Office of Solid Waste and Emergency Response, Technology Innovation Office.
2. Cookson, J.T., Jr., (1995). Bioremediation Engineering Design and Application. New York: McGraw-Hill, Inc.
3. King, R.B., Long, G.M., and Sheldon, J.K., (1998). Practical Environmental Bioremediation, Boca Raton, Florida: CRC Press.
4. Den Dooren De Jong, L. E.(1926). Thesis. University of Rotterdam, The Netherlands.
5. For a review of relevant research see Silver, S., Chakrabarty, A. M., Iglewski, B. and Kaplan, S., Pseudomonads. (1990). ASM. Washington, D.C.
This article originally appeared in the June 2002 issue of Environmental Protection, Vol. 13, No. 6, p. 43.
This article originally appeared in the 06/01/2002 issue of Environmental Protection.
About the Authors
Michael T. Saul, a geologist and vice president at CL Solutions, LLC.
Peter A. Vandenbergh, PhD, a microbiologist and vice president of science at Osprey Biotechnics, can be reached at by phone at 800.553.7785.